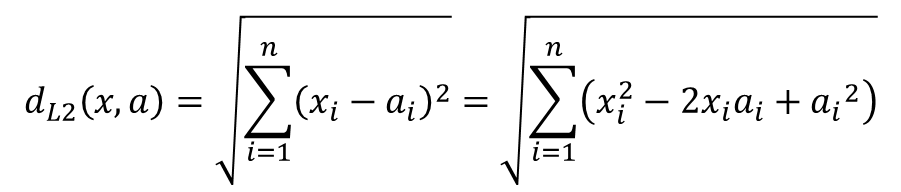Fast Euclidean Distance Calculation with Matlab Code
The Euclidean distance (also called the L2 distance) has many applications in machine learning, such as in K-Nearest Neighbor, K-Means Clustering, and the Gaussian kernel (which is used, for example, in Radial Basis Function Networks).
Calculating the Euclidean distance can be greatly accelerated by taking advantage of special instructions in PCs for performing matrix multiplications. Writing the Euclidean distance in terms of a matrix multiplication requires some re-working of the distance equation which we’ll work through below.
Using Squared Differences
The following is the equation for the Euclidean distance between two vectors, x and y.
Let’s see what the code looks like for calculating the Euclidean distance between a collection of input vectors in X (one per row) and a collection of ‘k’ models or cluster centers in C (also one per row).
The problem with this approach is that there’s no way to get rid of that for loop, iterating over each of the clusters. In the next section we’ll look at an approach that let’s us avoid the for-loop and perform a matrix multiplication instead.
Using Matrix Multiplication
If we simply expand the square term:
Then we can re-write our MATLAB code as follows (see the attached MATLAB script for a commented version of this).
XX = sum(X.^2, 2);
XC = X * C';
CC = sum(C.^2, 2)';
dists = sqrt(bsxfun(@plus, CC, bsxfun(@minus, XX, 2*XC)));No more for-loop! Because we are using linear algebra software here (MATLAB) that has been optimized for matrix multiplications, we will see a massive speed-up in this implementation over the sum-of-squared-differences approach.
MATLAB Example
I’ve uploaded a MATLAB script which generates 10,000 random vectors of length 256 and calculates the L2 distance between them and 1,000 models. Running in Octave on my Core i5 laptop, the sum-of-squared-differences approach takes about 50 seconds whereas the matrix multiplication approach takes about 2 seconds.
Fast Distance Comparisons
The above code gets you the actual Euclidean distance, but we can make some additional optimizations to it when we are only interested in _comparing _distances.
This occurs in K-Nearest Neighbor, where we are trying to find the ‘k’ data points in a large set which are closest to our input pattern. It also occurs in K-Means Clustering during the cluster assignment step, where we assign a data point to the closest cluster.
In these applications, we don’t need to know the actual L2 distance, we only need to compare distances–that is, determine which distance is smaller or larger.
The following equation expresses the comparison of the L2 distance between an input vector x _and two other vectors _a and b.
We will show that, in order to make this comparison, it is equivalent to instead compare the quantities:
The following figure shows the derivation of the above equivalence.
There is one additional modification we can make to this equation, which is to divide both sides by -2. This moves the 2 over to the pre-calculated term. Note that it also flips the comparison sign, so where we were previously looking for the minimum value, we are now looking for the maximum.